How to Create Different Tasks#
This notebook is a practical guide to the task-based architecture in spikeRNN. It shows how to work with built-in tasks and how to design your own.
First, make sure the spikeRNN package is installed
import sys
# !{sys.executable} -m pip install -e ..
Check all the available tasks
from rate.tasks import TaskFactory
available_tasks = TaskFactory.list_available_tasks()
print(f"Available tasks: {available_tasks}")
Available tasks: ['go_nogo', 'xor', 'mante']
Go-Nogo Task#
To create a Go-Nogo Task, you can directly use the task-based API:
from rate import GoNogoTask
settings = {'T': 200, 'stim_on': 20, 'stim_dur': 10, 'delay': 5}
task = GoNogoTask(settings)
stimulus, target, label = task.simulate_trial()
Or use the TaskFactory (where the Go-Nogo task is already instantiated)
# Set up task settings
settings = {'T': 200, 'stim_on': 20, 'stim_dur': 10, 'delay': 5}
# Create a GoNogo task
GoNogoTask = TaskFactory.create_task('go_nogo', settings)
stimulus, target, label = GoNogoTask.simulate_trial()
# Check the task type and label
print(f' {GoNogoTask}: {GoNogoTask.__class__.__name__} -> label: {label}')
<rate.tasks.GoNogoTask object at 0x71ef3357c8b0>: GoNogoTask -> label: 0
XOR Task#
Similarly, for XOR task, you can either directly use the task-based API:
from rate import XORTask
settings = {'T': 200, 'stim_on': 20, 'stim_dur': 10, 'delay': 5}
task = XORTask(settings)
stimulus, target, label = task.simulate_trial()
Or use TaskFactory (where XOR task is instantiated)
# Set up task settings
settings = {'T': 200, 'stim_on': 20, 'stim_dur': 10, 'delay': 5}
# Create a GoNogo task
XORTask = TaskFactory.create_task('xor', settings)
stimulus, target, label = XORTask.simulate_trial()
# Check the task type and label
print(f' {XORTask}: {XORTask.__class__.__name__} -> label: {label}')
<rate.tasks.XORTask object at 0x71ef348b5ba0>: XORTask -> label: diff
Mante Task#
from rate import ManteTask
settings = {'T': 200, 'stim_on': 20, 'stim_dur': 10, 'delay': 5}
task = ManteTask(settings)
stimulus, target, label = task.simulate_trial()
# Set up task settings
settings = {'T': 200, 'stim_on': 20, 'stim_dur': 10, 'delay': 5}
# Create a GoNogo task
ManteTask = TaskFactory.create_task('mante', settings)
stimulus, target, label = ManteTask.simulate_trial()
# Check the task type and label
print(f' {ManteTask}: {ManteTask.__class__.__name__} -> label: {label}')
<rate.tasks.ManteTask object at 0x71ef33587880>: ManteTask -> label: 1
Adding Custom Tasks#
Example 1: N-Back Working Memory Task#
Let’s create a more sophisticated custom task - an N-Back working memory task where the network must detect when the current stimulus matches the stimulus from N trials back.
from rate.tasks import AbstractTask
import numpy as np
class NBackTask(AbstractTask):
"""
N-Back working memory task.
The network must respond when the current stimulus matches
the stimulus from N trials back.
"""
def validate_settings(self):
required_keys = ['T', 'stim_on', 'stim_dur', 'n_back', 'n_stimuli']
for key in required_keys:
if key not in self.settings:
raise ValueError(f"Missing required setting: {key}")
# Validate timing
if self.settings['stim_on'] + self.settings['stim_dur'] >= self.settings['T']:
raise ValueError("Stimulus extends beyond trial duration")
# Validate n_back value
if self.settings['n_back'] < 1:
raise ValueError("n_back must be at least 1")
# Validate number of stimulus types
if self.settings['n_stimuli'] < 2:
raise ValueError("Must have at least 2 different stimuli")
def generate_stimulus(self, seed=False):
if seed:
np.random.seed(42)
T = self.settings['T']
stim_on = self.settings['stim_on']
stim_dur = self.settings['stim_dur']
n_stimuli = self.settings['n_stimuli']
# Initialize stimulus matrix (n_stimuli channels × time)
stimulus = np.zeros((n_stimuli, T))
# Choose a random stimulus type (0 to n_stimuli-1)
stim_type = np.random.randint(0, n_stimuli)
# Apply stimulus in the appropriate channel
stimulus[stim_type, stim_on:stim_on+stim_dur] = 1.0
return stimulus, stim_type
def generate_target(self, current_stim, seed=False, stimulus_history=None):
"""
Generate target based on current stimulus and history.
Args:
current_stim: Current stimulus type
seed: Random seed flag
stimulus_history: List of previous stimulus types
"""
T = self.settings['T']
stim_on = self.settings['stim_on']
stim_dur = self.settings['stim_dur']
n_back = self.settings['n_back']
# Initialize target (response after stimulus)
target = np.zeros(T-1)
response_start = stim_on + stim_dur
# Check if we have enough history and if current matches n-back
is_match = False
if stimulus_history and len(stimulus_history) >= n_back:
target_stimulus = stimulus_history[-n_back]
is_match = (current_stim == target_stimulus)
# Set target response
if is_match:
target[response_start:] = 1.0 # Respond for match
else:
target[response_start:] = 0.0 # No response for non-match
return target
def simulate_trial(self, seed=False, stimulus_history=None):
"""
Simulate a complete trial with history context.
"""
stimulus, stim_type = self.generate_stimulus(seed=seed)
target = self.generate_target(stim_type, seed=seed, stimulus_history=stimulus_history)
return stimulus, target, stim_type
# Test the N-Back task
settings = {
'T': 200,
'stim_on': 50,
'stim_dur': 30,
'n_back': 2, # 2-back task
'n_stimuli': 4 # 4 different stimulus types
}
nback_task = NBackTask(settings)
# Simulate a sequence of trials with history
stimulus_history = []
print("N-Back Task Simulation:")
print("=" * 50)
for trial in range(5):
stimulus, target, stim_type = nback_task.simulate_trial(stimulus_history=stimulus_history)
# Check if this is a match trial
is_match = False
if len(stimulus_history) >= settings['n_back']:
is_match = (stim_type == stimulus_history[-settings['n_back']])
print(f"Trial {trial+1}: Stimulus={stim_type}, Match={is_match}, Target_mean={target.mean():.1f}")
# Add to history
stimulus_history.append(stim_type)
print(f"\nStimulus history: {stimulus_history}")
print(f"Final stimulus shape: {stimulus.shape}")
print(f"Final target shape: {target.shape}")
N-Back Task Simulation:
==================================================
Trial 1: Stimulus=3, Match=False, Target_mean=0.0
Trial 2: Stimulus=3, Match=False, Target_mean=0.0
Trial 3: Stimulus=0, Match=False, Target_mean=0.0
Trial 4: Stimulus=0, Match=False, Target_mean=0.0
Trial 5: Stimulus=3, Match=False, Target_mean=0.0
Stimulus history: [3, 3, 0, 0, 3]
Final stimulus shape: (4, 200)
Final target shape: (199,)
Example 2: Delayed Match-to-Sample Task#
Another classic cognitive task where the network must remember a sample stimulus and then respond when a test stimulus matches it after a delay.
class DelayedMatchToSampleTask(AbstractTask):
"""
Delayed Match-to-Sample task.
Timeline:
1. Sample stimulus presentation
2. Delay period (working memory)
3. Test stimulus presentation
4. Response period (match/no-match decision)
"""
def validate_settings(self):
required_keys = ['T', 'sample_on', 'sample_dur', 'delay_dur', 'test_on', 'test_dur', 'n_stimuli']
for key in required_keys:
if key not in self.settings:
raise ValueError(f"Missing required setting: {key}")
# Validate timing sequence
sample_end = self.settings['sample_on'] + self.settings['sample_dur']
test_start = self.settings['test_on']
test_end = test_start + self.settings['test_dur']
if sample_end > test_start:
raise ValueError("Sample stimulus overlaps with test stimulus")
if test_end >= self.settings['T']:
raise ValueError("Test stimulus extends beyond trial duration")
if (test_start - sample_end) < self.settings['delay_dur']:
raise ValueError("Insufficient delay period")
def generate_stimulus(self, seed=False):
if seed:
np.random.seed(42)
T = self.settings['T']
sample_on = self.settings['sample_on']
sample_dur = self.settings['sample_dur']
test_on = self.settings['test_on']
test_dur = self.settings['test_dur']
n_stimuli = self.settings['n_stimuli']
# Initialize stimulus matrix (n_stimuli channels × time)
stimulus = np.zeros((n_stimuli, T))
# Generate sample stimulus
sample_type = np.random.randint(0, n_stimuli)
stimulus[sample_type, sample_on:sample_on+sample_dur] = 1.0
# Generate test stimulus (50% chance of match)
if np.random.rand() < 0.5:
test_type = sample_type # Match trial
is_match = True
else:
# Non-match trial - choose different stimulus
available_stimuli = [i for i in range(n_stimuli) if i != sample_type]
test_type = np.random.choice(available_stimuli)
is_match = False
stimulus[test_type, test_on:test_on+test_dur] = 1.0
return stimulus, {'sample': sample_type, 'test': test_type, 'match': is_match}
def generate_target(self, label, seed=False):
T = self.settings['T']
test_on = self.settings['test_on']
test_dur = self.settings['test_dur']
# Response starts after test stimulus
response_start = test_on + test_dur
target = np.zeros(T-1)
# Set target based on match/no-match
if label['match']:
target[response_start:] = 1.0 # Respond for match
else:
target[response_start:] = 0.0 # No response for non-match
return target
# Test the Delayed Match-to-Sample task
dms_settings = {
'T': 300,
'sample_on': 30,
'sample_dur': 20,
'delay_dur': 50,
'test_on': 120,
'test_dur': 20,
'n_stimuli': 3
}
dms_task = DelayedMatchToSampleTask(dms_settings)
print("Delayed Match-to-Sample Task:")
print("=" * 40)
# Run several trials
for trial in range(3):
stimulus, target, label = dms_task.simulate_trial(seed=True if trial==0 else False)
print(f"Trial {trial+1}:")
print(f" Sample: {label['sample']}, Test: {label['test']}")
print(f" Match: {label['match']}, Target response: {target.mean():.1f}")
print(f" Stimulus shape: {stimulus.shape}")
print()
Delayed Match-to-Sample Task:
========================================
Trial 1:
Sample: 2, Test: 0
Match: False, Target response: 0.0
Stimulus shape: (3, 300)
Trial 2:
Sample: 2, Test: 0
Match: False, Target response: 0.0
Stimulus shape: (3, 300)
Trial 3:
Sample: 2, Test: 2
Match: True, Target response: 0.5
Stimulus shape: (3, 300)
Example 3: Registering Custom Tasks with TaskFactory#
You can also register your custom tasks with the TaskFactory for easy access:
# Register custom tasks with TaskFactory
TaskFactory.register_task('nback', NBackTask)
TaskFactory.register_task('dms', DelayedMatchToSampleTask)
print("Available tasks after registration:")
print(TaskFactory.list_available_tasks())
# Now you can create tasks using the factory
nback_task_factory = TaskFactory.create_task('nback', {
'T': 200, 'stim_on': 50, 'stim_dur': 30, 'n_back': 2, 'n_stimuli': 4
})
dms_task_factory = TaskFactory.create_task('dms', {
'T': 300, 'sample_on': 30, 'sample_dur': 20, 'delay_dur': 50,
'test_on': 120, 'test_dur': 20, 'n_stimuli': 3
})
print(f"\nCreated {nback_task_factory.__class__.__name__} via factory")
print(f"Created {dms_task_factory.__class__.__name__} via factory")
Available tasks after registration:
['go_nogo', 'xor', 'mante', 'nback', 'dms']
Created NBackTask via factory
Created DelayedMatchToSampleTask via factory
Visualization: Understanding Task Structure#
Let’s visualize the stimulus and target patterns for our custom tasks to better understand their structure:
import numpy as np
import matplotlib.pyplot as plt
def plot_task_trial(task, title, max_tries=25, **kwargs):
"""Plot stimulus channels and target with annotated task events.
Tries multiple trials (max_tries) to obtain a non-flat target when needed.
Aligns target (T-1) to stimulus timeline (T).
"""
stimulus = target = label = None
for _ in range(max_tries):
stimulus, target, label = task.simulate_trial(**kwargs)
if np.any(target > 0):
break
T = stimulus.shape[1]
fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(12, 8), sharex=True)
# Plot stimulus channels with offsets
for i in range(stimulus.shape[0]):
ax1.plot(np.arange(T), stimulus[i, :] + i * 1.2, label=f'Channel {i}')
# Annotate known timing windows based on settings keys
st = getattr(task, 'settings', {})
if all(k in st for k in ['stim_on', 'stim_dur']):
ax1.axvspan(st['stim_on'], st['stim_on'] + st['stim_dur'], color='C7', alpha=0.2, label='Stimulus')
if all(k in st for k in ['sample_on', 'sample_dur']):
ax1.axvspan(st['sample_on'], st['sample_on'] + st['sample_dur'], color='C2', alpha=0.2, label='Sample')
if all(k in st for k in ['test_on', 'test_dur']):
ax1.axvspan(st['test_on'], st['test_on'] + st['test_dur'], color='C3', alpha=0.2, label='Test')
ax1.set_title(f'{title} - Stimulus Channels')
ax1.set_ylabel('Channel + Offset')
ax1.grid(True, alpha=0.3)
ax1.legend(loc='upper right', ncol=2)
# Plot target aligned to stimulus timeline
ax2.plot(np.arange(T - 1), target, 'r-', linewidth=2, label='Target Response')
ax2.set_title(f'{title} - Target Response (aligned)')
ax2.set_ylabel('Target Value')
ax2.set_xlabel('Time Steps')
ax2.grid(True, alpha=0.3)
ax2.set_ylim(-0.1, 1.1)
ax2.set_xlim(0, T - 1)
ax2.legend(loc='upper right')
# Consistency checks and helpful annotations
note_lines = []
# N-Back style label: integer channel
if isinstance(label, (int, np.integer)) and all(k in st for k in ['stim_on', 'stim_dur']):
active = np.where(stimulus[:, st['stim_on']:st['stim_on'] + st['stim_dur']].sum(axis=1) > 0)[0]
note_lines.append(f'Stimulus channel(s) active: {active.tolist()} | Label: {int(label)}')
if int(label) not in active.tolist():
note_lines.append('WARNING: label may not match active stimulus channel(s).')
# DMS label: dict with sample/test/match
if isinstance(label, dict) and {'sample', 'test', 'match'} <= set(label.keys()):
note_lines.append(f"Sample={label['sample']}, Test={label['test']}, Match={label['match']}")
if note_lines:
fig.text(0.02, 0.01, '\n'.join(note_lines), fontsize=10, family='monospace')
plt.tight_layout(rect=[0, 0.04, 1, 1])
plt.show()
return label
# Plot N-Back task trial (try until a response appears by re-sampling)
print("N-Back Task Visualization:")
nback_label = plot_task_trial(nback_task, "2-Back Task", stimulus_history=[0, 0], max_tries=100)
print(f"Label or info: {nback_label}")
print()
N-Back Task Visualization:
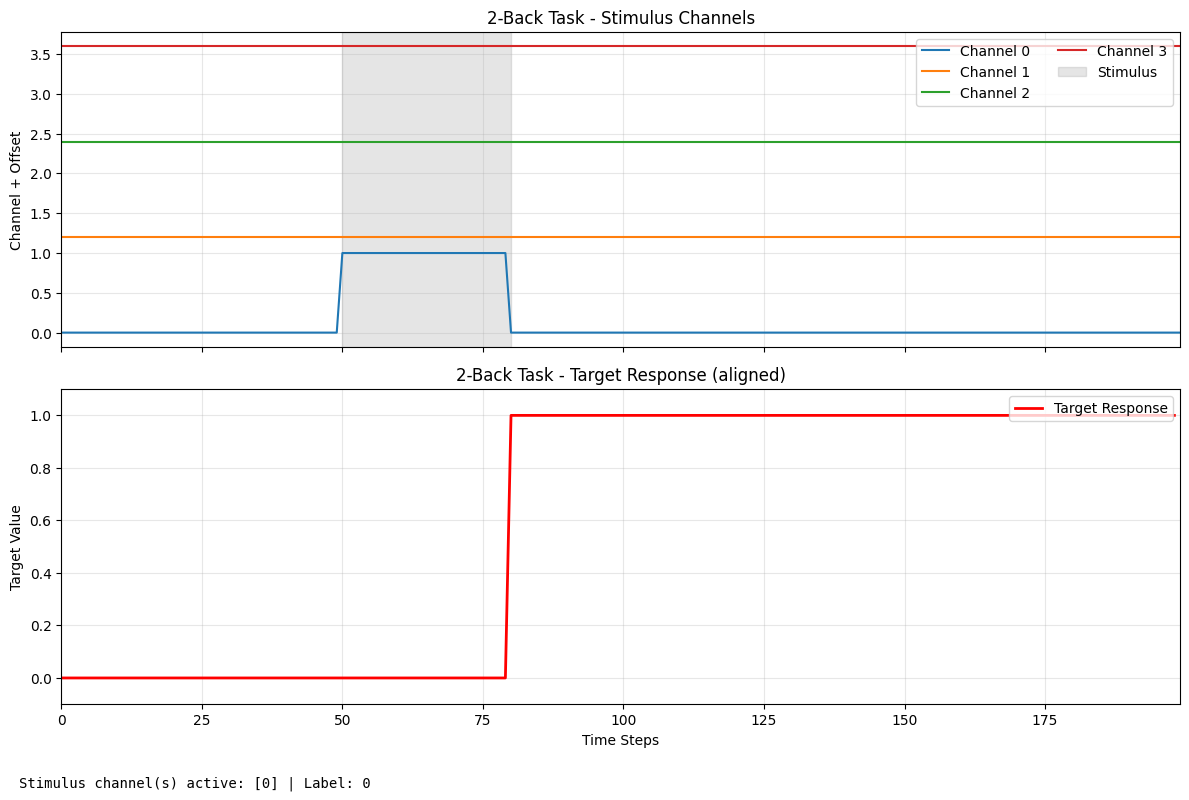
Label or info: 0
# Plot Delayed Match-to-Sample task trial
print("Delayed Match-to-Sample Task Visualization:")
dms_label = plot_task_trial(dms_task, "Delayed Match-to-Sample Task", seed=True)
print(f"Trial info: {dms_label}")
print()
Delayed Match-to-Sample Task Visualization:
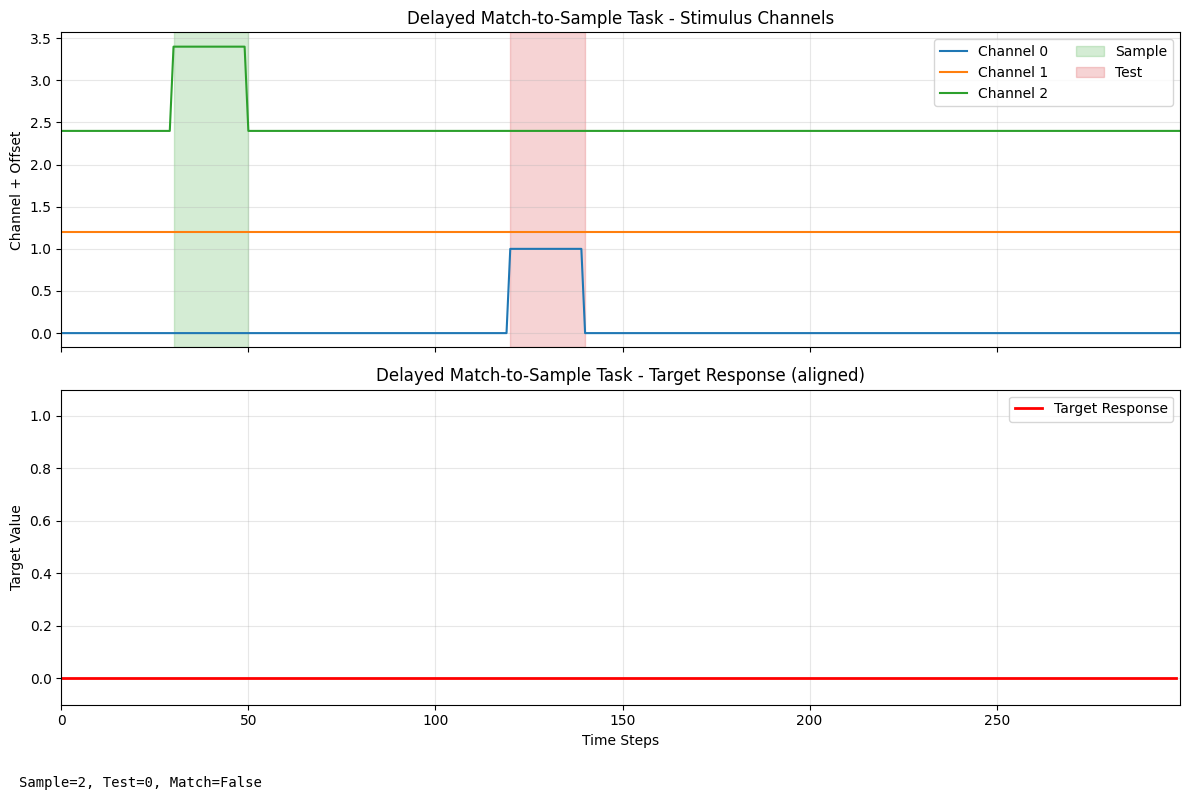
Trial info: {'sample': 2, 'test': np.int64(0), 'match': False}
Key Design Principles for Custom Tasks#
When creating custom tasks, follow these important principles:
Inherit from AbstractTask: Always inherit from
AbstractTaskto ensure compatibilityImplement required methods:
validate_settings(): Check that all required parameters are present and validgenerate_stimulus(): Create the input stimulus patterngenerate_target(): Create the target output pattern
Use descriptive settings: Make task parameters configurable through the settings dictionary
Handle edge cases: Validate timing, stimulus ranges, and other constraints
Document your task: Include clear docstrings explaining the task logic
Test thoroughly: Verify that your task generates expected patterns
Optional: Advanced Features#
You can also override the default simulate_trial() method if you need custom logic for combining stimulus and target generation, as shown in the N-Back example where we need to track stimulus history.

